-
Categories
-
Tags
3-gene TB score 3DQ laboratory aarhus university Aarthi Chary Academia academic community academic medicine acute myeloid addiction adipose tissue adolescents adult Adverse Events adversity africa aging AI AI in Healthcare AI-Assisted Hospital AIDS air pollution aldehyde metabolism alice milivinti ALL all department meeting all staff meeting allergies allergy allocation Allyship alyce adams AML amyloid analytics anesthesia Angioplasty annual retreat antagonism anthrax anti-hunger antibiotics antibodies antifungal antimicrobial antirac antiracism antiretroviral Antiretroviral Therapy antiviral Aortic Disorders Aortic Stenosis aortitis applied medicine applied physics arterial hypertension arthritis Artificial Intelligence ashkan afshin asian americans asthma at risk Atrial Fibrillation austin autoimmune diseases Azithromycin babies babies in uganda back pain bacteremia bacteria bacteriophages Bangladesh barbecue bartonella bbq bedside medicine bedside teaching behavior behavioral behavioral science benefit concert BeWell bias big data billing Biobanks biochemistry Biodesign bioengineering Bioethics bioinformatics Biomakers biomarker Biomedical Data biomedical ethics biomedical informatics biomedical model Biomedical Research Biomedical science biomedical workplace biomedicine biopharmaceuticals Biospecimen biotechnology bioterrorism birth cohort black and hispanic black lives matter blackburn blood and marrow transplantation blood drive bls blume lecture BMT body bollyky bone bone fractures bone health Bone scans born in bradford botulism brain gut connection Brain Health Brain Injury brain scans Breast Cancer breast cancer outcomes breast feeding Breast Oncology brian brian blackburn bubble budgeting bugs burnout bv“Genomic Landscape of Uveal Melanoma”“Genomic Landscape of Uveal Melanoma” c diff c difficile canary center Cancer cancer care cancer cells Cancer data cancer epidemiology cancer outcomes Cancer Pree cancer prevention cancer research cancer screening Cancer stage Information cancer treatment Cancer treatment needs candidiasis cannabis CAPA Cardiac Care Cardiology Cardiovascular Cardiovascular Disease Cardiovascular Institute cardiovascular medicine Cardiovascular Research cardiovascular surgery care care management career paths Careers CareEverywhere platform case case presentation case studies cases causes cdc cdh cdm dataset cec best practices CEDAR Project cell biology Cell Immune Therapy Cell Immune Tolerance Cell Rearch census census bureau census data center for asian health research and education center for digital health Center for Health Policy Center for Innovation in Global Health Center for Primary Care and Outcomes Research centre for applied education research cerc cesar vargas nunez Change changing etiology chemical and social exposure chicago chief fellows chief residents child development services child health childhood health childhood malnutrition children children's health China chronic diseases chronic fatigue chronic illness cigarettes circulatory support claims claims data Claims-Medical-Rx-Confinements clerkship climate change clinformatics clinic clinical Clinical Case Presentation clinical cases Clinical Data Clinical decision support clinical exam Clinical excellence clinical informatics clinical manifestations clinical protocol Clinical Providers wellbeing Clinical Quality Clinical Research Clinical Research Operations clinical skills clinical studies clinical teaching skills clinical trial design Clinical trials Clinicians clubhouse CME CMV cocci Coccidioidomycosis Coccidioidoycosis cognitive abilities Collecting large data collective trauma colon cancer columbia university combat injury common communication community Community Engagement Community Health community mental health community practice compensation Compliance Computational Genomics computer science Computer-interpretable Guidelines Concussion conflict in medicine conflict of interest connected health consult consultative medicine consumer health care contemplation by design continuing medical education Continuity Continuity of Care controlled trials Coronary artery disease Coronary Disease coronary stenting county public health coverage analysis COVID covid-19 covid19 creativity in medicine credence trial crispr Critical Care crohn's disease CROI culture Culture Change cushing disease CVI Cybele Renault cystic fibrosis cystric fybrosis Cytogenetics dana rose garfin danish populations data data center data quality data resources data science data sets Data sharing data training Data Warehouse datacommons datamart datasets date of death dean dean winslow deaths of despair decision making defense DEI demography dengue denied medical claims department wide event Dermatology design development Device Development diabetes diabetes research diabetes technology diabetic monitoring diagnoses diagnosis diagnostics dialysis Digestive Disease Clinical Conference digestive health digital health digital media digital phenotype digital technologies digitized health discovery discussion disease Disease States diseases disparities dissident doctor Diversity diversity week DNA DOD/SES/ZIPS dom DOM DEI domain-specific donor dora ho dosing drug Drug Combination Challenge Drug Development drug discovery Drug efficacy drug safety Drug Synergy drugs duration DVT early detection easter medicine Ebola ebv ecg interpretation ECHO ECMO economic development economic equity economics Education education outcomes education talks educators efficient health care delivery EHR EHR data EHR technology EHR-Based Studies elderly care electrocardiographic tracings Electronic Data Electronic Documents electronic health information Electronic Health Records electronic medical records Electronic Medical Reocrds electronic phenotyping Electronic Records elliot m tucker-drob ELSI emi lesure emotion employment end of life endocarditis endocrine oncology endocrinology endovascular engineering ENT environment environment-gene interaction environmental environmental health environmental influences EPIC epidemic epidemiological data epidemiology epigentics equity equity in medicine eRA Commons esophagus ethical legal and social implications ethics ethiopia etiologic studies evolution extremely large databases eye faculty faculty development faculty event faculty events faculty forum faculty happy hour faculty meeting faculty networking failure family id fatty liver fbi FDA FDA Inspection FDA Regulations febrile febrile neutropenia federal statistical research fellows didactic fellowship program fever fireside chat first 1000 days flukes fmig food allergies Food Systems Informatics foodborne illness forgive for good frederic luskin frontiers in diabetes fsrdc fungal Fungus gastric cancer gastroenterology gastroenterology and hepatology gastroesophagael reflex disease GD2 CAR T-cell Therapy gen silent gender gender dichotomies gender norm gender variables gene expression gene therapy General GI genetic genetic architecture genetic control genetic disease genetic origins genetic variants Genetics genetics and environment genome genome-wide data genomes genomic data Genomics GenoScan genotype genotypes geographic medicine Geriatrics gi GI Oncology GI Research Conference Gina Suh glenn steele global aging global burden of disease Global Health global health economics Global Oncology GMD Good Clinical Practice (GCP) google google street view vcars gram negative gram positive grand aides Grand Roun Grand Round Grand Rounds grant writing grants guatemala guide gun safety GWAS hacking healthcare handwashing HCT health health and disparities Health and Wellness Health Behaviors health care Health care information Health care insurance Health care markets health care outcomes health care systems Health data Health Disparities health educational systems Health Equity Health Information technology health initiative Health IT health metrics health mobility health outcomes health plus plus health policies health policy health related data health research health research and policy health sciences health statistics health transformation alliance Healthcare Healthcare database Healthcare information healthcare innovation healthcare IT Healthcare knowledge graph healthcare research healthcare research & quality healthcare workers Healthy Diet heart heart failure heart health heatlh policy help center hemapoetic Hematology Hematopoiesis hepatitis hepatitis c hepatology herpes herpes simplex Heterogeneous public data hewlett award hhv HIE high blood pressure high-risk Histone proteins history of medicine HIV HIV Journal Club holiday holiday party hormones hospitalist hosts howard hu hsv human human development human development survey Human genome Human Gut human mind Human Subjects Protection hypertension hypoxic IBD ICDS ichs ID ID Grand Rounds ID journal ID Lecture Series IDSA IDWeek ild illness imaging immigration immune Immune system immunity immunocompromised immunodeficiency Immunology immunomodulators immunosuppresion immunosuppression Immunotherapy inclusion inclusion 2020 inclusion 2021 inclusion 2022 inclusion 2023 inclusion 2024 inclusion rounds inclusive workplace inclusivity IncRNAs india inequities infecious diseases infection infections Infectious Diseaes infectious disease infectious Diseases Infectious GI infertility inflammation Influenza Informatics information technology Informed Consent innovative institutional approval insulin resistance intergenerational Internal Medicine internet industry internet of things internet saathi interoperability interstitial lung disease intra-abdominal ipums irritable bowel disease ISEXS jack rowe jackie ferguson james faghmous jason fletcher jerry reaven memorial jo robinson john groopman john w rowe joint Joint replacement joints jonathan fisher jonathan j king lecture jonathan kolstad Jonathan Li jose jose g montoya juneteenth junior faculty k award application kaiser permanente northern california kalanithi award kappagoda kayla kinsler keith wailoo kenetoplasts kidney Kidney Disease Kidney Stone Kidney Transplant kidneys lab lab-based research lambda language of functioning lauren gaydosh lawrence h kushi leadership leadership in healthcare learning collaborative Learning healthcare legal studies lesley park leukemia lgbtq+ life adversity line line infection line infections liver liver cancer liver inflammation long covid low returns lucy kalanithi lung cancer lung disease lupus Lupus Nephritis lyme disease lymphatic lymphoma machine learning machine-age tools MachineProse Malaria marcell alsan Marfan Syndrome marquitta white marriage patterens maternal health maternal health research MD MDS meaningful work medicaid medical medical apps medical care medical education Medical Image medical imaging medical practice medical technology Medical Terminology medicare Medicine medicine grand rounds Meditation meeting meeting with chief fellows Melanoma memorable presentations meningitis mental health mental health outcomes mental health services Mentorship meta analysis meta-data metabolic health methamphetamines methods methylation michael lindenmayer microaggressions microbes microbiology Microbiome Mindfulness minorities misdiagnoses MMSc mobile devices mobile medical apps modeling modern medicine molecular biology Molecular Genetics molecular imaging program Monitoring monoclonal montoya morbidity mortality movement MPNs mulit-ethnic birth cohort multi drug multi-disciplinary multi-omics mycobacterial disease Myeloma myocarditis narrative in medicine national center for health statistics National Disability Employment Awareness Month national immigration national longitudinal study National Lung Screening Trial national registry natural killer cells natural killers Natural language processing nba NCBI NCDs nchs neha barjatya nematodes neourology Nephrology networking Neurobiology neurodegeneration neurology neuromodulation neuroplasticity Neuroscience neurosciences Neurosurgery neutropenia new england jouranl of medicine newborn survival next generation nicole bush NIH NIH Clinical Center nipah nlp nodules Non-compliance Non-Human Subjects Research noninvasive NSCLC nuclear medicine nutrition nutrition science oakridge national laboratory ORNL obesity obesity management Observational Health Sciences and Informatics obstetrics and gynecology Oncology oncore Ontologies ophthalmology opioid opioid epidemic opportunistic infections opportunities Optimism optum optum data organ organ transplant Organ Transplantation ORSL orthopaedics orthopedics Osteoarthritis Osteoblast osteomyelitis osteoporosis otolaryngology outcomes P.Ananadan pacific islander health Pain Management Pain Medicine pain relief Palliative Care pandemic paradox parasites parasitic parent-child pairs parkinsons pathogens Pathology Pathophysiology pathways patient Patient Adherence Patient Care Patient Education patient engagement Patient Safety patient value patients Patietn Saety paul payer claims data pcha-uha pcp PCPH Pediatric Nephrology Pediatric Oncology pediatrics Peptides performance art PERFUSE perioperative care personalized health technology Personalized Medicine PGS phage-bacterial collaboration pharmaceutical Pharmacogenomics pharmacy PHATE PhDs Phenotype Phil Grant Philip Grant philipp koellinger phind PHIND seminar phs data phs trainees phsyiology phylogenetics Physical Examination physical health physician burnout Physician wellbeing physician-hospital integration physiology Placebo Placebo Effect plague plasticity pneumonia podcast policies policy policy intervention policy makers policy researchers policymakers pollutants population population based health Population Health population health sciences populations post docs post treatment Postoperative Outcomes PPCM practice-based research precision health precision medicine preclinical work prediction Predictive Models pregnancy pregnant premature death prenatal Prescription drugs presence preventative care Preventative Medicine prevention Prevention Research prevention studies Preventive Cardiology preventive care pride pride study Primary Care primary care and population health procedure codes products professional stress project baseline project brave heart promotion promotions prophylaxis Prostate cancer prostate gland prostatitis prosthetic provider tables Psychiatric Emergencies Psychiatric First aid Psychiatry psychological responses Psychology PTSD pubic safety Public Health Publication models publish Pulmonary pulmonary and critical care pulmonary critical care Pulmonary Diseases Pulmonary Fibrosis Pulmonary Health Pulmonary Hypertension quality care Quality Improvement Quality Incidents Quality Management quality oncology care quantitative imaging r2g2 racial disparities racial/ethnic minority Radiogenomics Radiology Radiology notes Radiomics rahul panicker randomize evaluations randomized controlled trial rare diseases rash reappointment recipients recovery trial Recruitment redwood city refugee health Regenerative Medicine remote learning reproductive medicine research research and infrastructure Research Human Subjects research tips researchers residency residency research program resilience Resistance resistant respirator respiratory Responsibilities returning rheumatic diseases rheumatology rhonda mcclinton-brown RIP risk factors risky behaviors robert shafer rubenstein lecture rv guha sam gambhir sampling sandro galea sanitation santa clara county santa rosa community health Sarcoma sccr school of medicine scientific writing secondary education self-care self-insured seminar SEPI sepsis septic sequencing serious illness services investment severe sex sex and gender sex differences sexual harassment sexually transmitted sgm shanthi simulation modeling skin skin/soft tissue sleep sleep health Sleep Medicine slow medicine smallpox smart sanitation social social challenges social determinants social genomics social media social mobility social services societal aging socioculture socioeconomic soft tissue spatial scales spectroscopy spectrum Spinal Conditions spine issues Spirituality Sports Medicine SPRC staff staff event staff meeting stanford 25 stanford cancer institute stanford center for clinical research stanford diabetes research center Stanford-China staph staph aureus Staphylococcal Infection startup startx state of department state of the department statistical learning statistical research statistics STD stem cells stephanie leonard stewardship STI stress stress test Stroke structural racism stuck at home concert students study study budgets Study Design suicide prevention superior canal dehiscence supplements Surgery suzan carmichael sven van egmond swelling Symposium synapse systemic Systems systems biology tata trusts teaching teaching medicine technology telemedicine temperature testosterone theranostics therapy thoracic Thoracic Oncology tick borne ticks tina hernandez-boussard Tobacco and Drug Use toxoplasmosis trainees training trans-disciplinary Transcriptional Data transgender health translational translational oncology translational research translational researcha transplant Transplant Immunology transplantation trauma Travel travelers traveling treatment trematodes trial management Trial Master File tropical diseases Tropical Medicine troponin truven Tuberculosis tularemia tumor type 2 diabetes ucla undiagnosed unique data sets university of texas unknown origin updates urban urinary tract infection Urologic Malignancy UTI vacation vaccination vaccine vaccines Valley Fever vanderbilt university variants Vascular Physiology vasculitis Veterans victoria udalova viral Viral Threats Virology virtual virtual data virtual reality virus Viruses vulnerable populations vzv wadhwani ai Wadhwani Institute wall street water water related wearable technologies wearable technology wearables Web Ontology Language Webinar weight management Wellbeing wellmd wellness western medicine when breath becomes air whsdm WikiDoc wildfires willem h ouwehand winslow women in medicine women in science and engineering women's education women's faculty luncheon women's faculty networking women's health work life balance work purpose working groups world bank World Health Organization worms writing a cv Yoga youth zika zoom tips
David Maslove, MD, MS
Assistant Professor
Department of Medicine and Critical Care
Queens University
ABSTRACT:
The diseases treated in the Intensive Care Units (ICU) are syndromic in nature, largely defined by a number of vague or arbitrary criteria. As a result, there exists significant case mixing within ICU syndromes, whereby different syndrome subtypes – or endotypes – are all treated the same way. This limits the effectiveness of evidence-based practices evaluated in large clinical trials, and makes critical care particularly suitable to a precision approach. Further enabling this approach is the incredible abundance of data generated in the ICU, upon which distinctions between patients can be made. Making the most of these data requires that they be systematically collected, vetted, merged, and analyzed, ideally in a near real-time environment that respects the fast-paced nature of ICU practice. We will explore some key challenges to this approach, with a focus on specific examples involving physiologic, clinical, and genomic data generated from critically ill and injured patients.

Teri Klein
Member of Academic Council
Professor (Research) of Biomedical Data Science and of Medicine (BMIR)
Stanford University
Abstract:
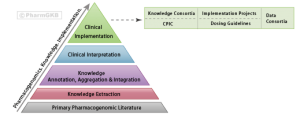
Pharmacogenomics (PGx) focuses on the use of genomic information to guide drug therapy and is a central component of precision medicine. Despite substantial progress in understanding how genetic variations impact drug efficacy and toxicity, the adoption of pharmacogenomics in clinical practice has been relatively slow. Major challenges in the implementation of pharmacogenomics knowledge include lack of awareness of the available evidence, unsure of how to interpret and use the genetic information, and lack of clear guidance on how to deliver information to the practitioners and patients. A central repository of pharmacogneomic knowledge is critical in addressing all of these challenges. The Pharmacogenomics Knowledgebase (PharmGKB) is a publically available premiere repository that collects, curates, and disseminates information about the impact of human genetic variation on drug responses. Through our research efforts and collaborations with pharmacogenomics research and clinical communities, we provide a comprehensive catalogue of genes and genetic variations that are most important for drug response phenotypes. I will describe the core content of our knowledgebase and discuss how we use the knowledge to support clinical implementation of PGx. In addition, I will highlight our collaboration with the Clinical Pharmacogenetics Implementation Consortium (CPIC) to develop freely available, peer-reviewed gene/drug practice guidelines for physicians that aids implementation of pharmacogenetic testing and improves the precision of drug selection and dosing. Lastly, I will present the development of The Pharmacogenomics Clinical Annotation Tool (PharmCAT), a software tool that extract all CPIC level-A variants from a genetic dataset (represented as a vcf), interpret the variant alleles, and generate a report that can then be used to inform prescribing decisions
References
- Whirl-Carrillo, E.M. McDonagh, J. M. Hebert, L. Gong, K. Sangkuhl, C.F. Thorn, R.B. Altman and T.E. Klein. “Pharmacogenomics Knowledge for Personalized Medicine”. Clinical Pharmacology & Therapeutics (2012) 92(4): 414-417.
- V. Relling, T.E. Klein. “CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network.” Clin Pharmacol Ther. 2011 Mar;89(3):464-7